Assay of Clinical Product Long-Term Delivery Systems for PLGA Properties
J. Garner1, S. Skidmore1, H. Park1, K. Park1, S. Choi2, Y. Wang2
1Akina, Inc. West Lafayette, IN 47906 USA
2Food and Drug Administration, Silver Spring, MD 20993, USA
Introduction
- Poly(lactide-co-glycolide) (PLGA) is a biodegradable polymer used in a wide variety of clinical products due to its capacity to biodegrade by hydrolysis into non-toxic lactic and glycolic acids.
- There are many different types of PLGA depending on the lactide:glycolide (LA:GA) ratio, endcap, and molecular weight.
- There is no good method established for assaying the PLGA component properties of microparticles used in injectable depot formulations, such as Risperdal® Consta® and Trelstar®.
- Such an assay is necessary for quality control as well as ensuring that proposed generic formulations provide qualitative and quantitative (Q1/Q2) sameness in regards to the reference product.
Purpose of this work is to establish a testing protocol which extracts PLGA from clinically used microparticle formulations and assays it to ensure Q1/Q2 compliance for parental depot formulations.
Methods
- Commercially purchased Risperdal Consta, which is a monthly injection, as well as Trelstar 3.75, 11.25, and 22.5 mg doses (1, 3, and 6 month injections, respectively) were dissolved in dichloromethane (DCM) (Fig. 1)
- Solutions filtered and dialyzed for three days (MWCO 6000-8000Da) against organic solvent.
- Subsequently, these solutions were concentrated, and precipitated in a stirring excess of hexane (Fig. 2) and dried under deep vacuum.
- The PLGA was then analyzed by gel permeation chromatography (GPC) (Fig. 3), 1H nuclear magnetic resonance (NMR) and 13C NMR (Fig. 4). (1)
 Figure 1.
Figure 1. Dissolution of microparticles
 Figure 2.
Figure 2. Precipitation in hexane
 Figure 3.
Figure 3. Waters Breeze 2 GPC system
 Figure 4.
Figure 4. NMR system Bruker AVIII-500HD
2
Results
- GPC was used to measure the molecular weight (Mol. Wt.) of PLGAs extracted from formulations (Fig. 5). These results processed against polystyrene standards for number average/weight average molecular weights.
- Fig. 6 shows example HNMR spectra and peak assignments.
- The LA:GA ratio was determined by relative peak integration at 5.2 ppm (LA, 1H) and 4.8 ppm (GA, 2H), respectively.
- 13C NMR was performed using cryoprobe for a total of 12,000 scans acquired over 12.5 hours to maximize signal/noise ratio.
- Peak at 14 ppm (red arrow in Fig. 7) correlates to alkyl endcap carbon and is indicative of ester endcap. Lack of peak indicates acid endcap. All data is summarized in Table 1.
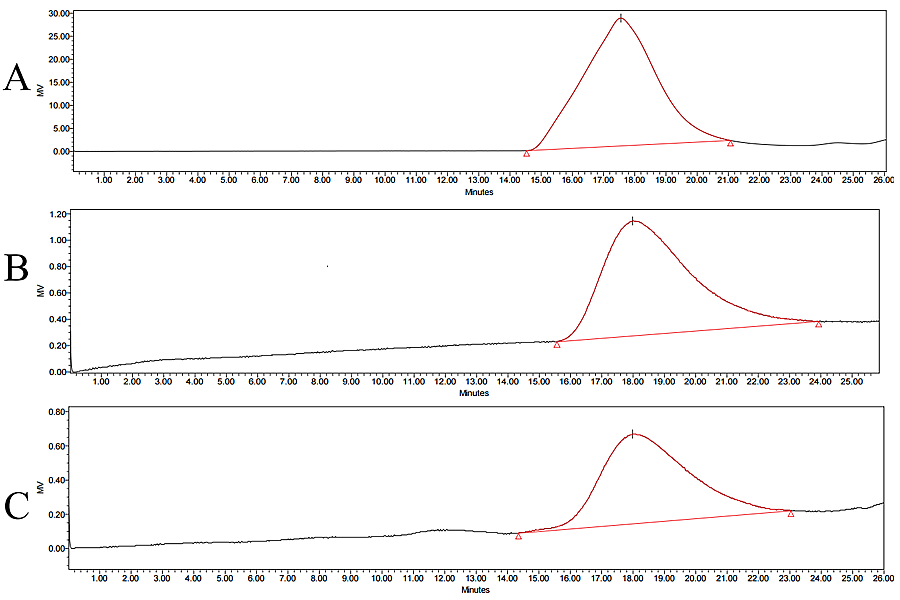 Figure 5.
Figure 5. Chromatograms of PLGA extracted from Risperdal Consta (A), Trelstar 11.25 mg (B), and Trelstar 22.5 mg (C).
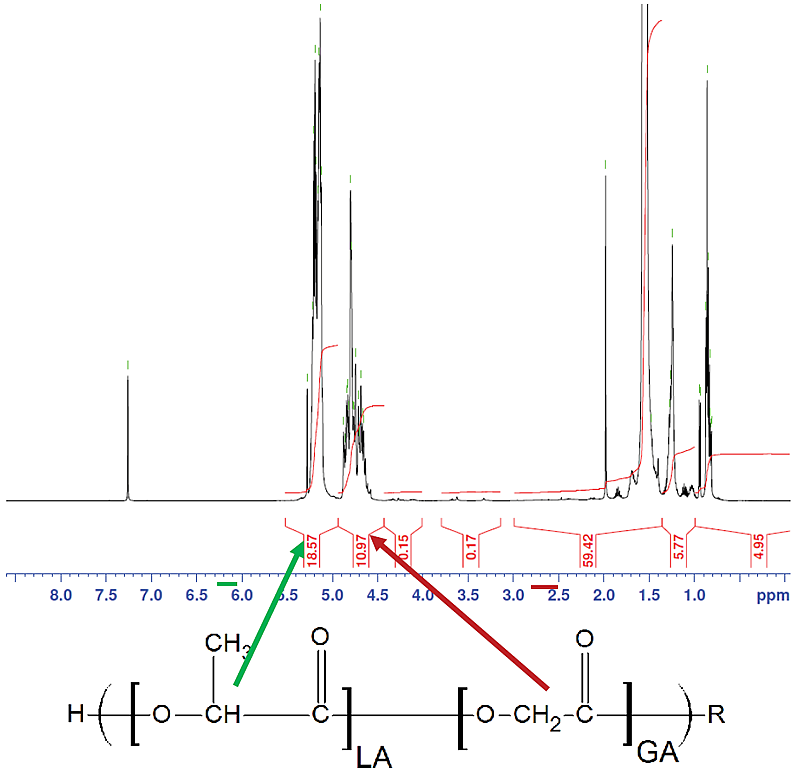 Figure 6.
Figure 6. Trelstar 22.5 PLGA H
1NMR
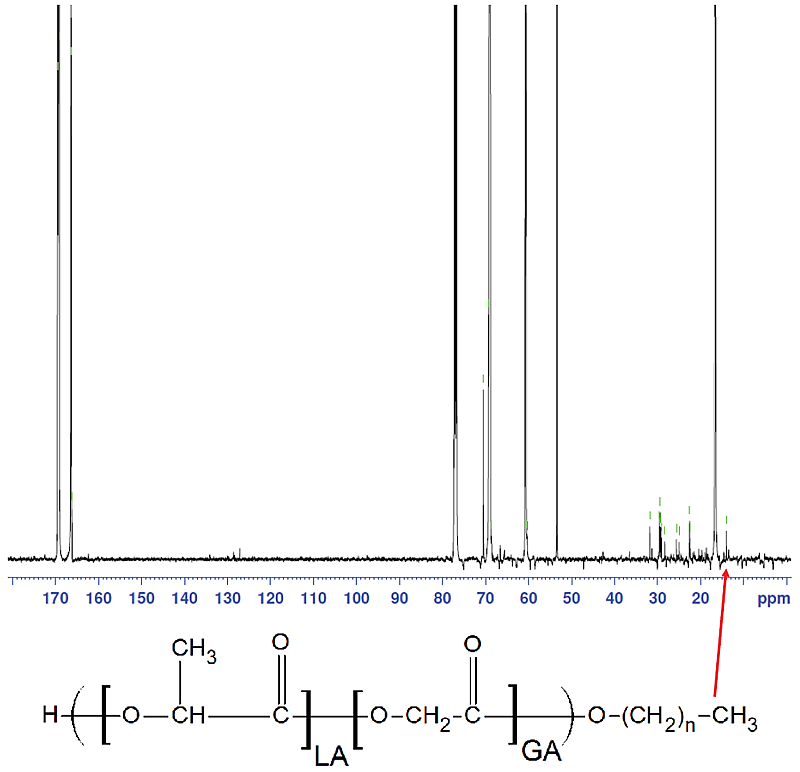 Figure 7.
Figure 7. Risperdal PLGA
13C NMR
| Product |
LA:GA ratio (molar) |
Mol. Wt. (Number average) |
Mol. Wt. (Weight average) |
End cap |
| Risperdal Consta |
78:22:00 |
44,875 |
111,142 |
Ester |
| Trelstar (3.75mg) |
52:48:00 |
25,192 |
85,207 |
Ester |
| Trelstar (11.25mg) |
74:26:00 |
47,214 |
72,286 |
Acid |
| Trelstar (22.5mg) |
77:23:00 |
46,368 |
74,042 |
N/A |
Table 1. Formulation PLGA parameters
Conclusion
- PLGAs were successfully extracted from formulations for assays of their parameters.
- Using the described method, results indicate that similar PLGAs were used for 3-month and 6-month Trelstar formulations. This is unexpected because the two have very different drug release profiles, 3 months vs 6 months.
- One limitation of the current assay method is that all PLGAs are extracted, regardless of identity, and assayed together. Thus, the assay presents average properties of different PLGAs rather than properties of individual PLGAs.
- The current method is suitable for formulations containing a single type of PLGA.
- There is a need to develop an advanced assay method which can separate individual PLGAs from a mixture of polymers in a single formulation.
Acknowledgements
This work is supported by Grant U01FD05168 from the Food and Drug Administration (FDA), Center for Drug Evaluation Research (CDER). The content is solely the responsibility of the authors and does not necessarily represent the official views of the Food and Drug Administration, Center for Drug Evaluation Research.
The authors would like to thank the Purdue Interdepartmental Nuclear Magnetic Resonance Facility (PINMRF) group (2). Supplemental data available at here.






